It’s easy to train a model against a particular dataset, but how does this model perform when introduced with new data? How do you know which machine learning model to use? Cross-validation answers these questions by assuring a model is producing accurate results and comparing those results against other models. Cross-validation goes beyond regular validation, the process of analyzing how a model does on its own training data, by evaluating how a model does on new data. Several different methods of cross-validation are discussed in the following sections:
The holdout cross-validation method involves removing a certain portion of the training data and using it as test data. The model is first trained against the training set, then asked to predict output from the testing set. This is the simplest form of cross-validation techniques, and is useful if you have a large amount of data or need to implement validation quickly and easily.

Typically the holdout method involves splitting a dataset into 20-30% test data and the rest as training data. These numbers can vary - a larger percentage of test data will make your model more prone to errors as it has less training experience, while a smaller percentage of test data may give your model an unwanted bias towards the training data. This lack of training or bias can lead to Underfitting/Overfitting of our model.
K-Fold Cross Validation helps remove these biases from your model by repeating the holdout method on k subsets of your dataset. With K-Fold Cross Validation, a dataset is broken up into several unique folds of test and training data. The holdout method is performed using each combination of data, and the results are averaged to find a total error estimation.
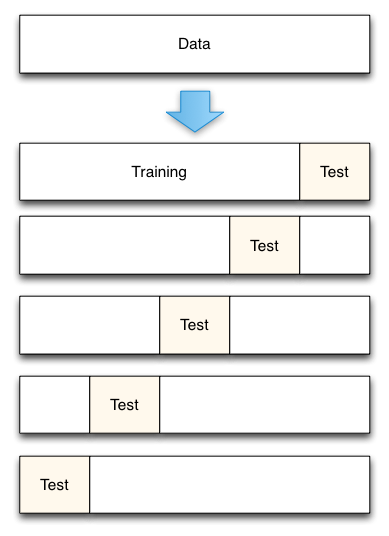
A “fold” here is a unique section of test data. For instance, if you have 100 data points and use 10 folds, each fold contains 10 test points. K-Fold Cross Validation is important because it allows you to use your complete dataset for both training and testing. It’s especially useful when evaluating a model using small or limited datasets.
Leave-P-Out Cross Validation (LPOCV) tests a model by using every possible combination of P test data points on a model. As a simple example, if you have 4 data points and use 2 test points, the model will be trained and tested as follows:
1: [ T T - - ] 2: [ T - T - ] 3: [ T - - T ] 4: [ - T T - ] 5: [ - T - T ] 6: [ - - T T ]
Where “T” is a test point, and “-” is a training point. Below is another visualization of LPOCV:
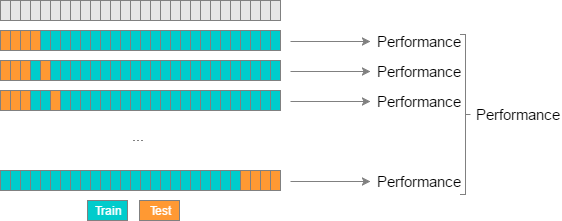
LPOCV can provide an extremely accurate error estimation, but can quickly become exhaustive for large datasets. The amount of testing iterations a model has to go through using LPOCV can be calculated using a mathematical combination n C P, with n being our total number of data points. We can see, for instance, that a LPOCV run using a dataset of 10 points with 3 test points would require 10 C 3 = 120 iterations.
Because of this, Leave-One-Out Cross Validation (LOOCV) is a commonly used cross-validation method. It is just a subset of LPOCV, with P being 1. This allows us to evaluate a model in the same number of steps as there are data points. LOOCV can also be seen as K-Fold Cross Validation, where the number of folds is equal to the number of data points.

Similar to K-Fold Cross Validation, LPOCV and LOOCV train a model using the full dataset. They are particularly useful when you’re working with a small dataset, but incur performance tradeoffs.
Cross-validation is a way to validate your model against new data. The most effective forms of cross-validation involve repeatedly testing a model against a dataset until every point or combination of points have been used to validate a model, though this comes with performance trade-offs. We discussed several methods of splitting a dataset for cross-validation:
There are many different types of machine learning models, including Linear/Logistic Regression, K-Nearest-Neighbors, and Support Vector Machines - but how do we know which type of model is the best for our dataset? Using a model unsuitable for our data will lead to less accurate predictions, and could lead to financial, physical, or other forms of harm. Individuals and companies should make sure to cross-validate any models they put into use.
The provided code shows how to split a set of data with the three discussed methods of cross-validation using Scikit-Learn, a Python machine learning library.
holdout.py splits a set of sample diabetes data using the Holdout Method. In scikit-learn, this is done using a function called train_test_split() which randomly splits a set of data into two portions:
TRAIN_SPLIT = 0.7 ... dataset = datasets.load_diabetes() ... x_train, x_test, y_train, y_test = train_test_split(...)
Note that you can change the portion of data used for training by changing the TRAIN_SPLIT value at the top. This should be a number from 0 to 1. Output from this file shows the number of training and test points used for the split. It may be beneficial to see the actual data points - if you would like to see these, uncomment the last two print statements in the script.
k-fold.py splits a set of data using the K-Fold Method. This is done by creating a KFold object initialized with the number of splits to use. Scikit-learn makes it easy to split data by calling KFold’s split() method:
NUM_SPLITS = 3 data = numpy.array([[1, 2], [3, 4], [5, 6], [7, 8], [9, 10], [11, 12]]) kfold = KFold(n_splits=NUM_SPLITS) split_data = kfold.split(data)
The return value of this is an array of train and test points. Note that you can play with the number of splits by changing the associated value at the top of the script. This script not only outputs the train/test data, but also outputs a nice bar where where you can track the progress of the current fold:
[ T T - - - - ] Train: (2: [5 6]) (3: [7 8]) (4: [ 9 10]) (5: [11 12]) Test: (0: [1 2]) (1: [3 4]) ...
leave-p-out.py splits a set of data using both the Leave-P-Out and Leave-One-Out Methods. This is done by creating LeavePOut/LeaveOneOut objects, the LPO initialized with the number of splits to use. Similar to KFold, the train-test data split is created with the split() method:
P_VAL = 2 data = numpy.array([[1, 2], [3, 4], [5, 6], [7, 8]]) loocv = LeaveOneOut() lpocv = LeavePOut(p=P_VAL) split_loocv = loocv.split(data) split_lpocv = lpocv.split(data)
Note that you can change the P value at the top of the script to see how different values operate.
|
# An example of the Holdout Cross-Validation split import pandas from sklearn import datasets from sklearn.model_selection import train_test_split # The percentage (as a decimal) of our data that will be training data TRAIN_SPLIT = 0.7 # The diabetes dataset contains the following columns: columns = [ 'age', 'sex', 'bmi', 'map', 'tc', 'ldl', 'hdl', 'tch', 'ltg', 'glu' ] # Load the diabetes dataset dataset = datasets.load_diabetes() print(dataset) # Create a pandas DataFrame from the diabetes dataset dataframe = pandas.DataFrame(dataset.data, columns=columns) # Split via the holdout method x_train, x_test, y_train, y_test = train_test_split( dataframe, dataset.target, train_size=TRAIN_SPLIT, test_size=1-TRAIN_SPLIT) print("""\ The holdout method removes a certain portion of the training data and uses it as test data. Ideally, the data points removed are random on each run. The following output shows a set of sample diabetes data split into test and training data: """) # Print our test and training data print("Total diabetes data points: {}".format(len(dataframe.index))) print("# of training data points: {} (~{}%)".format(len(x_train), TRAIN_SPLIT*100)) print("# of test data points: {} (~{}%)\n".format(len(x_test), (1-TRAIN_SPLIT)*100)) print("If you'd like to see the actual data points, uncomment the print statements at the bottom of this script.") # print("Training data:\n{}\n".format(x_train)) # print("Test data:\n{}".format(x_test)) |
# An example of K-Fold Cross Validation split
import numpy
from sklearn.model_selection import KFold
# Configurable constants
NUM_SPLITS = 3
# Create some data to perform K-Fold CV on
data = numpy.array([[1, 2], [3, 4], [5, 6], [7, 8], [9, 10], [11, 12]])
# Perform a K-Fold split and print results
kfold = KFold(n_splits=NUM_SPLITS)
split_data = kfold.split(data)
print("""\
The K-Fold method works by splitting off 'folds' of test data until every point has been used for testing.
The following output shows the result of splitting some sample data.
A bar displaying the current train-test split as well as the actual data points are displayed for each split.
In the bar, "-" is a training point and "T" is a test point.
""")
print("Data:\n{}\n".format(data))
print('K-Fold split (with n_splits = {}):\n'.format(NUM_SPLITS))
for train, test in split_data:
output_train = ''
output_test = ''
bar = ["-"] * (len(train) + len(test))
# Build our output for display from the resulting split
for i in train:
output_train = "{}({}: {}) ".format(output_train, i, data[i])
for i in test:
bar[i] = "T"
output_test = "{}({}: {}) ".format(output_test, i, data[i])
print("[ {} ]".format(" ".join(bar)))
print("Train: {}".format(output_train))
print("Test: {}\n".format(output_test))
|
# Example of LOOCV and LPOCV splitting import numpy from sklearn.model_selection import LeaveOneOut, LeavePOut # Configurable constants P_VAL = 2 def print_result(split_data): """ Prints the result of either a LPOCV or LOOCV operation Args: split_data: The resulting (train, test) split data """ for train, test in split_data: output_train = '' output_test = '' bar = ["-"] * (len(train) + len(test)) # Build our output for display from the resulting split for i in train: output_train = "{}({}: {}) ".format(output_train, i, data[i]) for i in test: bar[i] = "T" output_test = "{}({}: {}) ".format(output_test, i, data[i]) print("[ {} ]".format(" ".join(bar))) print("Train: {}".format(output_train)) print("Test: {}\n".format(output_test)) # Create some data to split with data = numpy.array([[1, 2], [3, 4], [5, 6], [7, 8]]) # Our two methods loocv = LeaveOneOut() lpocv = LeavePOut(p=P_VAL) split_loocv = loocv.split(data) split_lpocv = lpocv.split(data) print("""\ The Leave-P-Out method works by using every combination of P points as test data. The following output shows the result of splitting some sample data by Leave-One-Out and Leave-P-Out methods. A bar displaying the current train-test split as well as the actual data points are displayed for each split. In the bar, "-" is a training point and "T" is a test point. """) print("Data:\n{}\n".format(data)) print("Leave-One-Out:\n") print_result(split_loocv) print("Leave-P-Out (where p = {}):\n".format(P_VAL)) print_result(split_lpocv) |
1. https://towardsdatascience.com/cross-validation-in-machine-learning-72924a69872f
2. https://machinelearningmastery.com/k-fold-cross-validation/
3. https://www.quora.com/What-is-cross-validation-in-machine-learning
4. http://www.ebc.cat/2017/01/31/cross-validation-strategies/
Acknowledge: Python Machine Learning supervised by Amirsina Torfi, 2019.